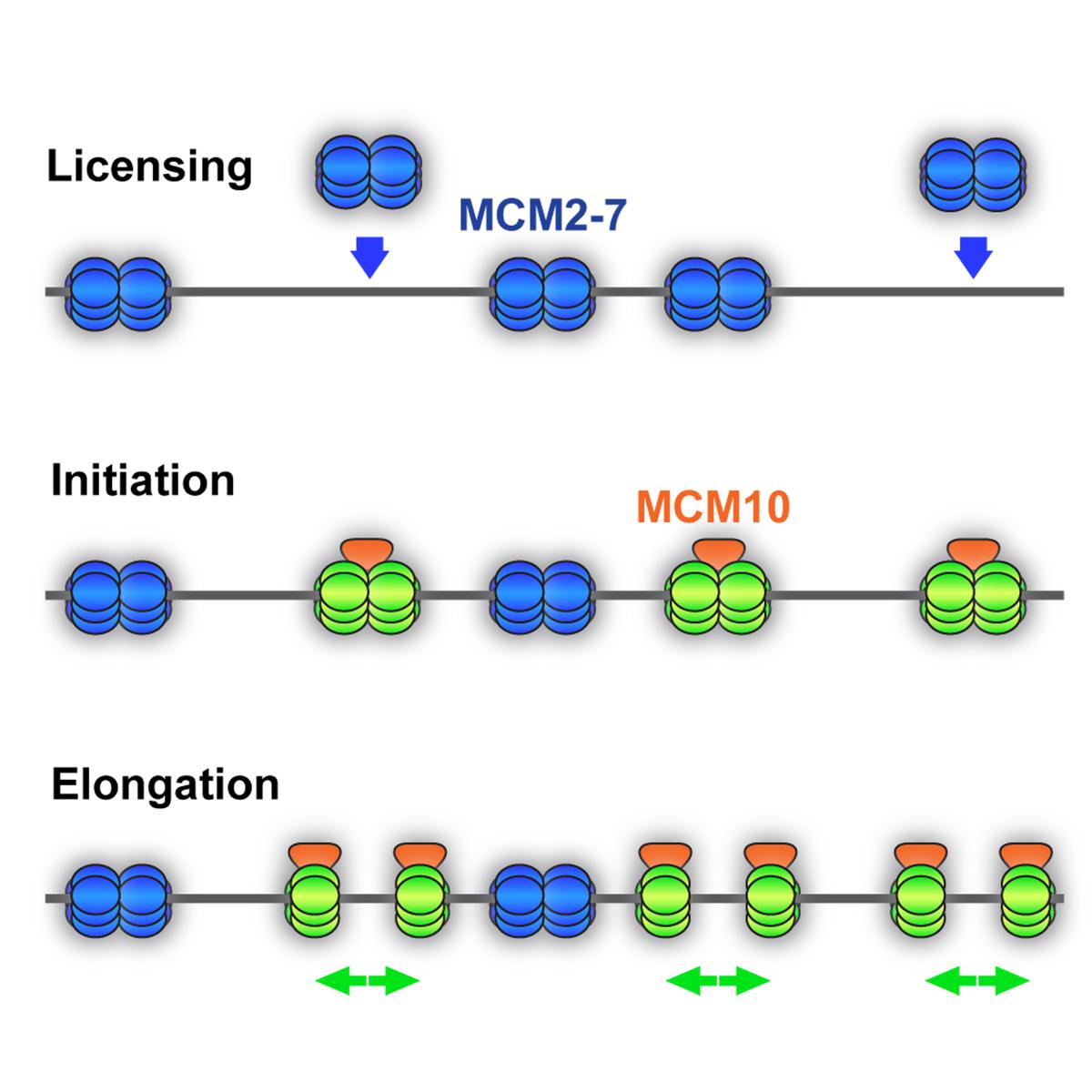
DNA replication takes place in S-phase of each cell cycle to duplicate the genome prior to cell division. This process begins with “licensing” thousands of chromosomal loci by loading the core machinery of the replicative helicase, composed of minichromosome maintenance proteins 2-7 (Mcm2-7), to establish origins of replication. Once a sufficiently high number of origins have been licensed, cells transition from G1- to S-phase of the cell cycle.
As S-phase begins, a subset of origins recruits protein factors to stimulate origin firing –the initiation of DNA synthesis. One factor essential to replication initiation is minichromosome maintenance protein 10 (Mcm10). The DNA replication machinery, or replisome, then begins DNA synthesis by two replication forks that move bidirectionally away from each origin. Mcm10 is also critical to maintain replisome stability and promote the complete duplication of genomic regions that are inherently hard-to-replicate. Unreplicated, single-stranded DNA causes “replication stress”.
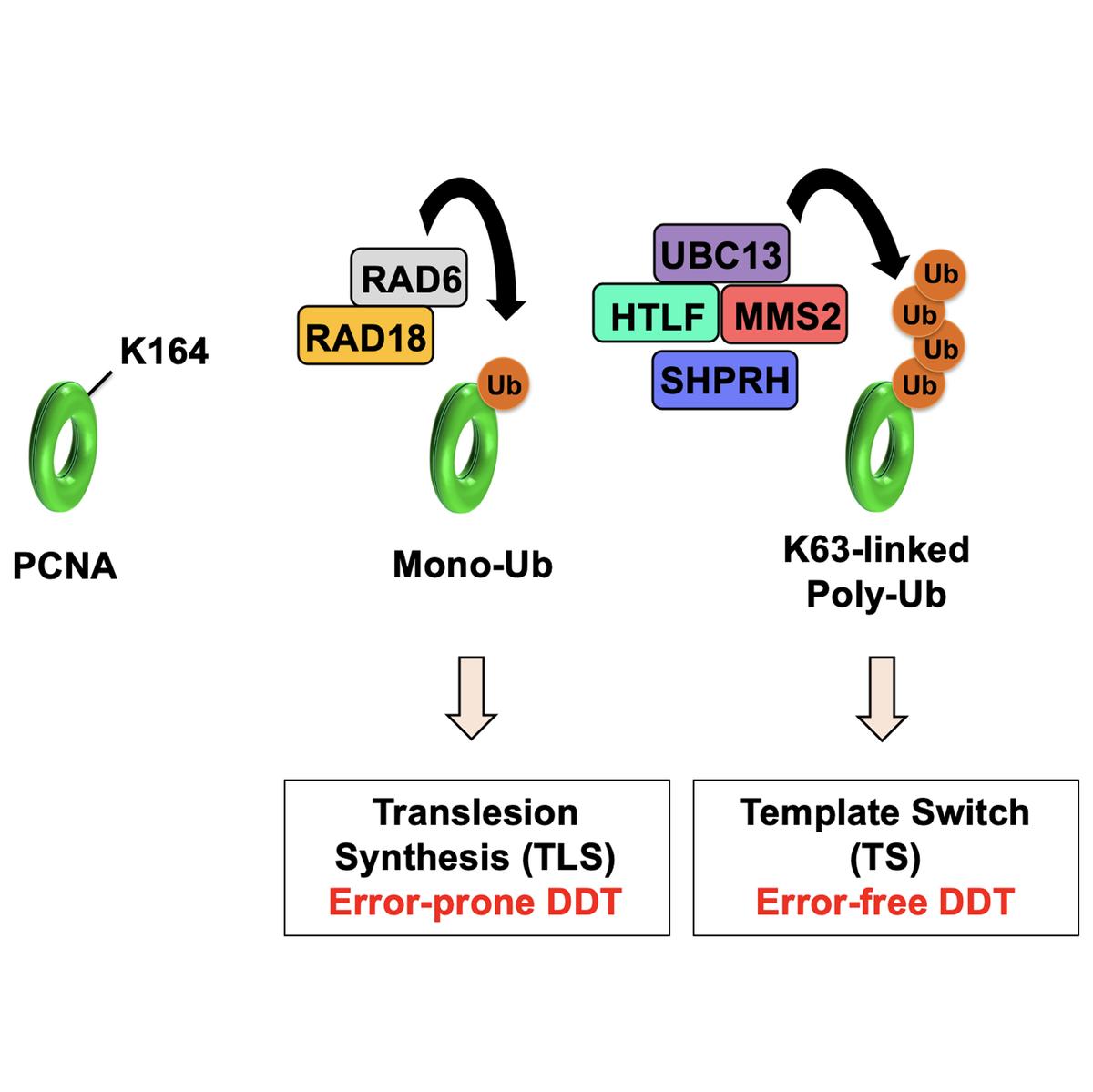
DNA synthesis also requires the replicative sliding clamp proliferating cell nuclear antigen (PCNA) that encircles DNA and tethers DNA polymerases to promote efficient replication. PCNA is critical to fill in unreplicated, single-stranded gaps in the DNA that cause a replication stress response. This pathway is regulated by the ubiquitination of the conserved lysine (K) 164 residue of PCNA. This pathway is called the DNA damage tolerance (DDT) pathway, because it is also used to replicate over-damaged DNA.
Another central player in the DDT pathway is the E3 ubiquitin ligase radiation sensitive 18 (Rad18). One conserved role of Rad18 and its binding partner Rad6 is the activation of DDT through the attachment of ubiquitin to PCNA. In addition to its role in the replication stress response, Rad18 also regulates repair of dsDNA breaks (DSBs), which are particularly toxic DNA lesions. Through its role in homologous recombination, Rad18 functions to ensure that DSBs are repaired via an error-free mechanism, thus preventing chromosomal rearrangements, including radial formations.
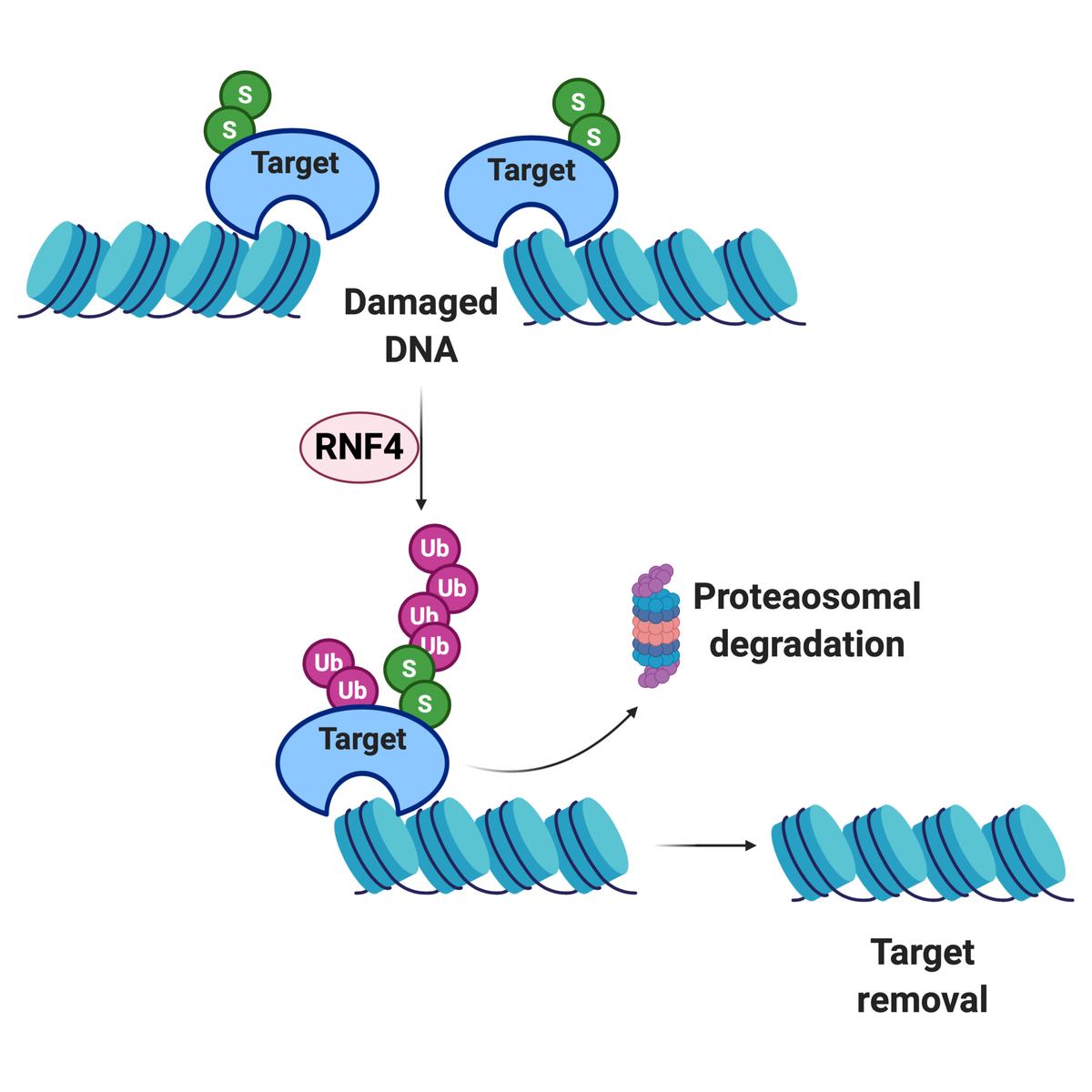
Cells that experience mild but chronic replication stress (e.g., due to MCM10 deficiency) require the RING finger protein 4 (RNF4) for survival. RNF4 is a small ubiquitin-like modifier (SUMO) targeted E3 ubiquitin ligase, or STUbL, and bridges the SUMOylation and ubiquitination signaling cascades during the replication stress response. RNF4 directly interacts with SUMO protein chains attached to target proteins to catalyze the addition of ubiquitin to mark proteins for degradation via the proteasome.
CRISPR-Cas9 technology enables chemical-genetic screening in human cell lines, allowing us to interrogate chemical-genetic interactions across a library of defined deletion mutants. In collaboration with the Myers Lab, we are developing chemical-genetic screens with a targeted library and computational analysis pipelines to predict chemical compound modes-of-action. Using these approaches, we can more efficiently identify candidate drug therapeutics and build a comprehensive drug library to help realize precision medicine.